Protein details/Phosphorylation: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
|||
| Line 5: | Line 5: | ||
This section provides the following information: | This section provides the following information: | ||
*'''Source''' - | *'''Source''' - | ||
*'''Kinase Protein''' - | *'''Kinase Protein''' - | ||
*'''Kinase Gene''' - | *'''Kinase Gene''' - | ||
*'''Residue''' - | *'''Residue''' - | ||
*'''Notes''' - | *'''Notes''' - | ||
==Source of information== | ==Source of information== | ||
| Line 15: | Line 15: | ||
==Data access== | ==Data access== | ||
The collected data is processed and stored in '''[https://data.glygen.org/ data.glygen.org]''' in following datasets. | |||
Homo Sapiens (Human) Datasets | |||
* Human Phosphorylation Sites (UniProtKB; [https://data.glygen.org/GLY_000274 GLY_000274]) | |||
* Human Phosphorylation Sites (iPTMNet; [https://data.glygen.org/GLY_000519 GLY_000519]) | |||
Hepatitis C Virus Datasets | |||
* | |||
==Data harmonization== | ==Data harmonization== | ||
Revision as of 21:02, 19 November 2021
The Phosphorylation section of the Protein information page in GlyGen provides the detailed list of phosphorylation sites and kinase proteins responsible for phosphorylation.
Phosphorylation
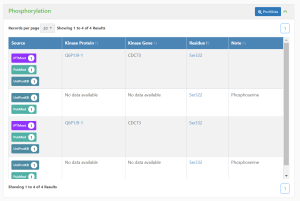
This section provides the following information:
- Source -
- Kinase Protein -
- Kinase Gene -
- Residue -
- Notes -
Source of information
The Phosphorylation data is collected and integrated from UniProtKB, PubMed, and iPTMnet databases.
Data access
The collected data is processed and stored in data.glygen.org in following datasets.
Homo Sapiens (Human) Datasets
- Human Phosphorylation Sites (UniProtKB; GLY_000274)
- Human Phosphorylation Sites (iPTMNet; GLY_000519)
Hepatitis C Virus Datasets
Data harmonization
Data filtering