Protein details/Single nucleotide variation
Jump to navigation
Jump to search
The Single Nucleotide Variation section of the Protein details page in GlyGen provides information on disease and non-disease associated mutations. These are most commonly nonsynonymous mutations which cause a different amino acid to be produced at a given position.
Single Nucleotide Variation
Single nucleotide variation is presented in 2 tabs on the Protein details page in GlyGen.
Disease associated Mutations
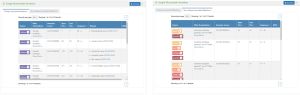
This tab shows a list of mutations in the genetic sequence that are associated with a disease. The following columns are presented in the table:
- Source - GlyGen evidence linking to the databases and papers that provided the glycosylation information
- Filter annotations - Pass/fail filter results from mapping cancer terms to Disease Ontology ID. Eg. Somatic mutation passed 1 out of 6 filters: patient freq. (2.0%)
- Genomic locus - The genomic position of the reported mutation. Eg. Chr7:81399287
- Start Pos. - Start position of the site in the amino acid sequence. Eg. 1
- End Pos. - End position of the site in the amino acid sequence. Eg. 1
- Sequence - Reference or wild-type amino acid residue change resulting from variation. Eg. M→ L
- Disease - DOID and term corresponding to reported disease. Eg. thyroid gland cancer (DOID:1781)
- MAF - The frequency at which the second most common allele occurs in a given population.
Non-disease associated Mutations
This tab shows a list of mutations in the genetic sequence that are not associated with a disease. The following columns are presented in the table:
- Source - GlyGen evidence linking to the databases and papers that provided the glycosylation information
- Filter annotations -
- Genomic locus -
- Start Pos. -
- End Pos. -
- Sequence -
- MAF -