Processed data file
The processed data file is used to upload the final data values of an glycan array experiment to the Glycan array data repository as part of the processed data upload. The file is an Excel table that may consist of multiple sheet. However one sheet has to be labeled "Repo Data" and has to follow the format instructions below. Only this sheet will be used to extract data from and load them to the repository. The sheet is designed to be human readable as well and can contain additional information which is ignored by the repository.
File Format
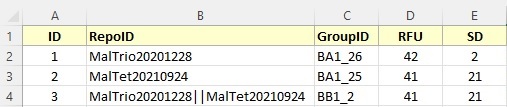
The processed data file is a standard Excel spreadsheet in *.xlsx format. The file may contain more than one sheet but one of the sheets need to be labeled "Repo Data". This sheet has to contain the following columns with the corresponding label in row one. All columns have to be in the spreadsheet. But columns labeled as optional might be left empty.
ID (required)
An user chosen ID that has to be unique for each row. This ID will not be imported to the repository but will be used by the repository to report errors in individual rows during data submission. The ID may be an auto-increment number or any alpha-numerical text.
RepoID (required)
ID of the feature in the repository (Internal ID). This ID must be already present in the repository and must be used in the slide that the processed data is associated with. If a row describes a mixture of features printed on the same spot the IDs are separated by "||" (e.g., Feature123||Feature124). Any alpha-numerical text is allowed as long as that feature ID is present in the repository.
GroupID (optional)
This column contains the group ID that is associated with the feature when creating the block layout. As long as all spots with the same feature/concentration are averaged into a single value this column can be left empty. However, if on the same slide a feature is present multiple times and the intensities are not average into one value (e.g., one value for all replicated of the feature on block 1 and another for block 2) the group ID is used to identify which feature this intensity value belongs to. Group IDs are alpha-numerical strings that need to match the corresponding group IDs in the slide/block layout.
RFU (required)
Averaged intensity value for the features. This value will be extracted by the repository and has to be a number or floating point number.
SD (required)
The standard deviation after averaging the intensity values of all spots with the same feature. This value will be extracted by the repository and has to be a number or floating point number.
Additional columns (optional)
Additional columns can be added to the end of the table. These columns will be ignored by the repository.
GLAD extension

To load data into the GLAD software an extended version of this format is used that contains additional columns beside the five columns described above.
Name (required)
Human readable name of the feature.
CV (required)
The coefficient of variation is calculated out of the standard deviation (SD) and average intensity value (RFU).
Rank (optional)
A user defined column that can be used to providing an additional sorting criteria beyond the RFU column.
Name2 (optional)
A column used to provide a secondary name or feature sequence. This column is not extracted by the repository.
Notes (optional)
A column to add user defined notes to each row.