Frequently Asked Questions: Difference between revisions
No edit summary |
|||
| (16 intermediate revisions by 3 users not shown) | |||
| Line 7: | Line 7: | ||
===How to Use Simple Protein Search?=== | ===How to Use Simple Protein Search?=== | ||
*You can search for proteins by specifying their UniProtKB Accession numbers, their specific structures or the specific biochemical contexts within which they are found. For details, please refer to https:// | *You can search for proteins by specifying their UniProtKB Accession numbers, their specific structures or the specific biochemical contexts within which they are found. For details, please refer to https://www.glygen.org/protein_search.html#tutorial | ||
===How to Use Simple Glycoprotein Search?=== | ===How to Use Simple Glycoprotein Search?=== | ||
*You can search for glycoproteins by specifying their UniProtKB Accession numbers, their specific structures or the specific biochemical contexts within which they are found. For details, please refer to https:// | *You can search for glycoproteins by specifying their UniProtKB Accession numbers, their specific structures or the specific biochemical contexts within which they are found. For details, please refer to https://www.glygen.org/glycoprotein_search.html#tutorial | ||
===What does protein-centric data include?=== | ===What does protein-centric data include?=== | ||
| Line 20: | Line 20: | ||
*UniProtKB canonical accession is an accession assigned to the protein isoform chosen to be the canonical sequence to which all positional annotation refers in the UniProtKB entry page. | *UniProtKB canonical accession is an accession assigned to the protein isoform chosen to be the canonical sequence to which all positional annotation refers in the UniProtKB entry page. | ||
*UniProtKB represents different isoforms of the same protein by assigning it a protein accession followed by the number of the isoform.For example: for protein accession P38398, different isoforms are represented as P38398-1, P38398-2, P38398-3, P38398-4, etc. where P38398-1 is the chosen canonical accession. | *UniProtKB represents different isoforms of the same protein by assigning it a protein accession followed by the number of the isoform.For example: for protein accession [https://www.uniprot.org/uniprot/P38398 P38398], different isoforms are represented as P38398-1, P38398-2, P38398-3, P38398-4, etc. where P38398-1 is the chosen canonical accession. | ||
*UniProtKB uses specific criteria for choosing the canonical sequence for the entry. To know more about canonical isoforms and the canonical choosing criteria, refer to the UniProtKB [https://www.uniprot.org/help/canonical_and_isoforms help] page. | *UniProtKB uses specific criteria for choosing the canonical sequence for the entry. To know more about canonical isoforms and the canonical choosing criteria, refer to the UniProtKB [https://www.uniprot.org/help/canonical_and_isoforms help] page. | ||
<div style="float: right;"> [[#top|[top]]]</div> | <div style="float: right;"> [[#top|[top]]]</div> | ||
| Line 63: | Line 63: | ||
*The policy of the [https://proconsortium.org/ Protein Ontology (PRO)] is to only represent proteoforms that have been experimentally observed. For example, if glycosylation is observed on two different sites, each of the singly modified forms can be represented in PRO; however, the proteoform with glycosylation on both sites will not be represented unless there is solid experimental evidence for its existence. In this way, the number of proteoforms per protein does not grow too large, and the forms represented are those that are of highest interest to biologists. | *The policy of the [https://proconsortium.org/ Protein Ontology (PRO)] is to only represent proteoforms that have been experimentally observed. For example, if glycosylation is observed on two different sites, each of the singly modified forms can be represented in PRO; however, the proteoform with glycosylation on both sites will not be represented unless there is solid experimental evidence for its existence. In this way, the number of proteoforms per protein does not grow too large, and the forms represented are those that are of highest interest to biologists. | ||
=== How can search for all proteins that bear my glycan/GlyTouCan Ac? === | ===How can search for all proteins that bear my glycan/GlyTouCan Ac?=== | ||
* Go to the '''Explore''' tab and click on '''Glycoprotein'''. | *Go to the '''Explore''' tab and click on '''Glycoprotein'''. | ||
* In the '''Advanced Search''', add your GlyTouCan accession in the '''Interacting Glycan''' field and click on '''Search.''' | *In the '''Advanced Search''', add your GlyTouCan accession in the '''Interacting Glycan''' field and click on '''Search.''' | ||
* Please refer to | *Please refer to [[Frequently Asked Questions#How do I find a GlyTouCan accession for my glycan composition or structure?|How do I find a GlyTouCan for my glycan structure]] on how to find a GlyTouCan accession for your desired glycan. | ||
<div style="float: right;"> [[#top|[top]]]</div> | <div style="float: right;"> [[#top|[top]]]</div> | ||
==Glycan FAQs== | ==Glycan FAQs== | ||
| Line 85: | Line 85: | ||
**Visit GlyGen's [https://www.glygen.org/glycan_search.html#composition_search composition search] to search for a glycan within the GlyGen set (please note: The GlyGen glycan set captures roughly ~30% of the entire GlyTouCan collection.) | **Visit GlyGen's [https://www.glygen.org/glycan_search.html#composition_search composition search] to search for a glycan within the GlyGen set (please note: The GlyGen glycan set captures roughly ~30% of the entire GlyTouCan collection.) | ||
**or utilize the [https://glycosmos.gitlab.io/compo2wurcsui/ compo2wurcs] tool developed by [https://glycosmos.org/ GlyCosmos] to search against the entire GlyTouCan database. | **or utilize the [https://glycosmos.gitlab.io/compo2wurcsui/ compo2wurcs] tool developed by [https://glycosmos.org/ GlyCosmos] to search against the entire GlyTouCan database. | ||
**If the composition is not available in GlyTouCan, please refer to FAQ [[Frequently Asked Questions#How can I register my glycan structure into GlyTouCan.3F| | **If the composition is not available in GlyTouCan, please refer to FAQ [[Frequently Asked Questions#How can I register my glycan structure into GlyTouCan.3F|How can I register my glycan structure into GlyTouCan?]] to see how to register your glycan composition. | ||
*If additional information (such as arrangement of monosaccharides, linkage positions, anomeric configuration, glycosidic bonds, etc) about the glycan is known: | *If additional information (such as arrangement of monosaccharides, linkage positions, anomeric configuration, glycosidic bonds, etc) about the glycan is known: | ||
**Visit GlyTouCan [https://glytoucan.org/Structures/graphical Graphic Input] | **Visit GlyTouCan [https://glytoucan.org/Structures/graphical Graphic Input] | ||
**Draw the glycan structure and provide available linkage, glycosidic bonds, etc information. | **Draw the glycan structure and provide available linkage, glycosidic bonds, etc information. | ||
**Click '''SEARCH''' | **Click '''SEARCH''' | ||
**If the drawn glycan structure is not present in GlyTouCan, please refer to the FAQ [[Frequently Asked Questions#How can I register my glycan structure into GlyTouCan.3F| | **If the drawn glycan structure is not present in GlyTouCan, please refer to the FAQ [[Frequently Asked Questions#How can I register my glycan structure into GlyTouCan.3F|How can I register my glycan structure into GlyTouCan?]] on how to register your glycan. | ||
*If a sequence/string-representation (such as GlycoCT, IUPAC extended,IUPAC condensed, KCF ,WURCS, etc) is known: | *If a sequence/string-representation (such as GlycoCT, IUPAC extended,IUPAC condensed, KCF ,WURCS, etc) is known: | ||
**Visit GlyCosmos [https://glycosmos.org/glytoucans Text Input] | **Visit GlyCosmos [https://glycosmos.org/glytoucans Text Input] | ||
**Insert the string format to get the accession. | **Insert the string format to get the accession. | ||
**To convert your sequence into a desired format, please refer to FAQ [[Frequently Asked Questions#How can I convert my glycan sequence | **To convert your sequence into a desired format, please refer to FAQ [[Frequently Asked Questions#How can I convert my glycan sequence/string-format to different desired formats (e.g IUPAC, WURCS, GlycoCT, LinearCode, etc.)?|How can I convert my glycan sequence/string-format to different desired formats (e.g IUPAC, WURCS, GlycoCT, LinearCode, etc.)?]] | ||
**If the GlyTouCan accession is not found, please refer to FAQ [[Frequently Asked Questions#How can I register my glycan structure into GlyTouCan.3F| | **If the GlyTouCan accession is not found, please refer to FAQ [[Frequently Asked Questions#How can I register my glycan structure into GlyTouCan.3F|How can I register my glycan structure into GlyTouCan?]] on how to register your sequence. | ||
===How can I register my glycan structure into GlyTouCan?=== | ===How can I register my glycan structure into GlyTouCan?=== | ||
| Line 127: | Line 127: | ||
*Go to the '''EXPLORE''' tab on the GlyGen home page and select '''Glycan Search'''. | *Go to the '''EXPLORE''' tab on the GlyGen home page and select '''Glycan Search'''. | ||
*Under '''Simple Search''', select Protein from '''Any category''' add your UniProtKB accession in the search box. | *Under '''Simple Search''', select Protein from '''Any category''' add your UniProtKB accession in the search box. | ||
*Click on '''Search'''. | *Click on '''Search'''. | ||
===How do I input the structure of the required glycan?=== | |||
You can use the '''[https://gnome.glyomics.org/restrictions/GNOme_GlyGen.browser.html GNOme]''' (beta testing is currently underway) widget to search for your desired glycan structure or composition. | |||
please see: https://gnome.glyomics.org/restrictions/GNOme_GlyGen.browser.html?GalNAc=1&Gal=1 | |||
By clicking on structures you can move further along glycan tree to retrieve a more specific structure (i.e with defined linkage information) | |||
for eg: https://gnome.glyomics.org/restrictions/GNOme_GlyGen.browser.html?saccharide=G16612QG | |||
You can right-click on your desired structure to see the glycan entry in GlyGen database, where you can retrieve additional information. (go to "Found Glycoprotein" section on the individual glycan entry page to find the associated proteins). | |||
Alternatively, you can use the GlyGen composition search to search for your glycan with desired monosaccharides composition. | |||
e.g.: https://www.glygen.org/glycan_list.html?id=8691cfa82cb8eea7bf6b68995ae37bc2 | |||
You can click on the desired GlyTouCan accession to retrieve more information about the glycan. | |||
Another option is to use the [https://glytoucan.org/Structures/graphical GlyTouCan's graphic input tool] to draw your desired glycan and retrieve the GlyTouCan accession. | |||
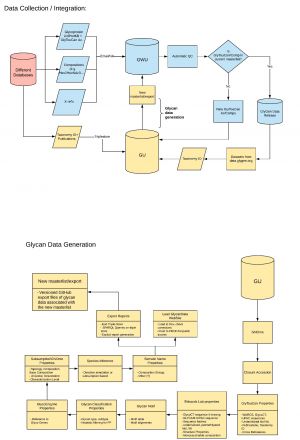
[[File:Glycan data flow 1.jpg|alt=|thumb|Glycan data flow]] | |||
Once you have the GlyTouCan accession you can use GlyGen's [https://www.glygen.org/glycan_search.html glycan] or [https://www.glygen.org/glycoprotein_search.html glycoprotein] searches to find your desired output. | |||
===How is glycan data collected, integrated, and processed in GlyGen?=== | |||
GlyGen processes glycoprotein-centric and glycan-centric data from different sources like GlyConnect, UniCarbKB, GlyTouCan, MatrixDB, etc in coordination between GlyGen team members at George Washington University and Georgetown University. GlyGen is also part of the [http://www.glyspace.org/ GlySpace] alliance and coordinates with team members [https://docs.google.com/document/d/1-ZJfHxDRHujKXImHkV2ytNWHBkZ3Ym16hLOJw3aCfxk/edit biocuration] activities. The overview of this process can be seen in image. | |||
==General FAQs== | ==General FAQs== | ||
===Does GlyGen have a tutorial on how to use the site?=== | ===Does GlyGen have a tutorial on how to use the site?=== | ||
* Tutorials for specific searches can be accessed through the '''EXPLORE''' tab on GlyGen home page. | *Tutorials for specific searches can be accessed through the '''EXPLORE''' tab on GlyGen home page. | ||
===What are the different resources integrated into GlyGen?=== | ===What are the different resources integrated into GlyGen?=== | ||
* Click [https://www.glygen.org/license.html here] to understand which resources are integrated into GlyGen. | *Click [https://www.glygen.org/license.html here] to understand which resources are integrated into GlyGen. | ||
===What licence does GlyGen use?=== | ===What licence does GlyGen use?=== | ||
* GlyGen’s data is licensed under CC BY 4.0. You are free to share (copy and redistribute the material in any medium or format) and adapt (remix, transform, and build upon the material) for any purpose, even commercially as long as you give appropriate credit and attribution. To learn about CC BY 4.0, click [https://creativecommons.org/licenses/by/4.0/ here]. | *GlyGen’s data is licensed under CC BY 4.0. You are free to share (copy and redistribute the material in any medium or format) and adapt (remix, transform, and build upon the material) for any purpose, even commercially as long as you give appropriate credit and attribution. To learn about CC BY 4.0, click [https://creativecommons.org/licenses/by/4.0/ here]. | ||
* GlyGen’s source code and data processing scripts are licenced under GNU General Public License v3.0 which gives the user freedom to use the software for any purpose, to change the software to suit the needs, to share the software with your friends and neighbors, and also the freedom to share the changes a user makes. To learn about GNU GPL V3.0, visit the GNU [https://www.gnu.org/licenses/gpl-3.0.en.html licenses] page. | *GlyGen’s source code and data processing scripts are licenced under GNU General Public License v3.0 which gives the user freedom to use the software for any purpose, to change the software to suit the needs, to share the software with your friends and neighbors, and also the freedom to share the changes a user makes. To learn about GNU GPL V3.0, visit the GNU [https://www.gnu.org/licenses/gpl-3.0.en.html licenses] page. | ||
<div style="float: right;"> [[#top|[top]]]</div> | <div style="float: right;"> [[#top|[top]]]</div> | ||
===What is GlyGen's policy regarding copyright and database distribution?=== | ===What is GlyGen's policy regarding copyright and database distribution?=== | ||
* You must give appropriate credit to GlyGen (by referring to GlyGen for the whole resource or Object ID for individual datasets) and may do so in any reasonable manner, but not in any way that suggests the licencor endorses you or your use. | *You must give appropriate credit to GlyGen (by referring to GlyGen for the whole resource or Object ID for individual datasets) and may do so in any reasonable manner, but not in any way that suggests the licencor endorses you or your use. | ||
* Datasets made available through GlyGen include archival of versions. We recommend authors to use Creative Commons Attribution licence (CC BY) for all versions when they share their datasets with GlyGen. | *Datasets made available through GlyGen include archival of versions. We recommend authors to use Creative Commons Attribution licence (CC BY) for all versions when they share their datasets with GlyGen. | ||
* This means that users of GlyGen are entitled to use, reproduce, disseminate or display these datasets provided the original authors and GlyGen are attributed. | *This means that users of GlyGen are entitled to use, reproduce, disseminate or display these datasets provided the original authors and GlyGen are attributed. | ||
===How can I access the GlyGen Data?=== | ===How can I access the GlyGen Data?=== | ||
* GlyGen data can be accessed through the '''DATA''' tab on the GlyGen homepage or directly via GlyGen [https://data.glygen.org/ data] page. Data can also be be accessed by querying SPARQL endpoint (coming in future) or through GlyGen Web Service [https://api.glygen.org/ API] | *GlyGen data can be accessed through the '''DATA''' tab on the GlyGen homepage or directly via GlyGen [https://data.glygen.org/ data] page. Data can also be be accessed by querying SPARQL endpoint (coming in future) or through GlyGen Web Service [https://api.glygen.org/ API] | ||
===How can I download GlyGen data?=== | ===How can I download GlyGen data?=== | ||
* ''To download a list of results (with one or more filters) or to download data specific to an individual record:'' | *''To download a list of results (with one or more filters) or to download data specific to an individual record:'' | ||
*Go to the '''EXPLORE''' tab on the GlyGen home page and select a category for your search (e.g. protein, glycan, glycoprotein). | *Go to the '''EXPLORE''' tab on the GlyGen home page and select a category for your search (e.g. protein, glycan, glycoprotein). | ||
| Line 166: | Line 195: | ||
**Go to the '''DATA''' tab on the GlyGen homepage. | **Go to the '''DATA''' tab on the GlyGen homepage. | ||
**Select '''view more''' on the desired data object and select the '''download''' button. | **Select '''view more''' on the desired data object and select the '''download''' button. | ||
**To know more about GlyGen data objects go to General FAQ | **To know more about GlyGen data objects go to General FAQ [[Frequently Asked Questions#What is a GlyGen data object?|What is a GlyGen data object]].<div style="float: right;"> [top]</div> | ||
===What is a GlyGen data object?=== | ===What is a GlyGen data object?=== | ||
* GlyGen data is organized into individual data objects which are assigned a unique GlyGen ID (e.g. [https://data.glygen.org/GLYDS000001 GLYDS000001]). | *GlyGen data is organized into individual data objects which are assigned a unique GlyGen ID (e.g. [https://data.glygen.org/GLYDS000001 GLYDS000001]). | ||
* Each GlyGen data object can be accessed through the '''DATA''' tab on the GlyGen home or directly via the Glygen [https://data.glygen.org/ data] page. | *Each GlyGen data object can be accessed through the '''DATA''' tab on the GlyGen home or directly via the Glygen [https://data.glygen.org/ data] page. | ||
* Each data object has a README describing the source, contributors, integration process and quality control workflow, etc. Go to General FAQ 8 for more information. | *Each data object has a README describing the source, contributors, integration process and quality control workflow, etc. Go to General FAQ 8 for more information. | ||
* The data objects can be filtered based on different categories (Protein, Proteoform, Glycan), by species (Homo sapiens, Mus musculus, Rattis norvegicus etc.), and by file type (.csv, .fasta, .png, etc.). | *The data objects can be filtered based on different categories (Protein, Proteoform, Glycan), by species (Homo sapiens, Mus musculus, Rattis norvegicus, SARS-CoV-1, SARS-CoV-2 etc.), and by file type (.csv, .fasta, .png, etc.). | ||
<div style="float: right;"> [[#top|[top]]]</div> | <div style="float: right;"> [[#top|[top]]]</div> | ||
===What is the format of the README files?=== | ===What is the format of the README files?=== | ||
* All readme files follow the BioCompute specifications. Technical specifications can be found [https://github.com/biocompute-objects/BCO_Specification here] . | *All readme files follow the BioCompute specifications. Technical specifications can be found [https://github.com/biocompute-objects/BCO_Specification here] . | ||
[[File:General faq21.png|515x515px|alt=|thumb|Data integration process in GlyGen. ]] | [[File:General faq21.png|515x515px|alt=|thumb|Data integration process in GlyGen. ]] | ||
| Line 186: | Line 215: | ||
===How can I submit my data?=== | ===How can I submit my data?=== | ||
* Please use our Contact [https://www.glygen.org/contact.html page] to submit your data to GlyGen. | *Please use our Contact [https://www.glygen.org/contact.html page] to submit your data to GlyGen. | ||
===How often is the GlyGen data updated?=== | ===How often is the GlyGen data updated?=== | ||
* GlyGen data is released every six months during March-April and August-September of each year. | *GlyGen data is released every six months during March-April and August-September of each year. | ||
* Minor updates to the data are made when a bug or a data issue has been found and addressed. | *Minor updates to the data are made when a bug or a data issue has been found and addressed. | ||
===How can I access the previous versions of the data?=== | ===How can I access the previous versions of the data?=== | ||
* Go to the '''DATA''' tab on the GlyGen home page. | *Go to the '''DATA''' tab on the GlyGen home page. | ||
*Click '''view details''' on the desired data object. | *Click '''view details''' on the desired data object. | ||
| Line 202: | Line 231: | ||
===Which UniProtKB and GlyTouCan accessions are inclded in GlyGen?=== | ===Which UniProtKB and GlyTouCan accessions are inclded in GlyGen?=== | ||
* GlyGen maintains a strict protein (UniProtKB) and glycan (GlyTouCan) accession list compiled based on different criteria. | *GlyGen maintains a strict protein (UniProtKB) and glycan (GlyTouCan) accession list compiled based on different criteria. | ||
* The UniProtKB accession list serves as a reference for generating protein-centric and proteoform-centric datasets, whereas the GlyTouCan accession list is used for generating glycan-centric datasets. The criteria and the statistics may be subject to change with every major and minor release. | *The UniProtKB accession list serves as a reference for generating protein-centric and proteoform-centric datasets, whereas the GlyTouCan accession list is used for generating glycan-centric datasets. The criteria and the statistics may be subject to change with every major and minor release. | ||
* '''Protein:''' | *'''Protein:''' | ||
**Please see the following species specific GlyGen datasets to see the current GlyGen protein masterlists: | |||
** Please see the following species specific GlyGen datasets to see the current GlyGen protein masterlists: | ***Homo sapiens [TaxID:9606]: [https://data.glygen.org/GLYDS000001 GLYDS000001] | ||
*** Homo sapiens [TaxID:9606]: [https://data.glygen.org/GLYDS000001 GLYDS000001] | ***Mus musculus [TaxID:10090]: [https://data.glygen.org/GLYDS000007 GLYDS000007] | ||
*** Mus musculus [TaxID:10090]: [https://data.glygen.org/GLYDS000007 GLYDS000007] | ***Rattus norvegicus [TaxID:10116]:[https://data.glygen.org/GLYDS000244 GLYDS000244] | ||
*** Rattus norvegicus [TaxID:10116]:[https://data.glygen.org/GLYDS000244 GLYDS000244] | ***Hepatitis C virus (genotype 1a, isolate H) [TaxID:11108]: [https://data.glygen.org/GLYDS000344 GLYDS000344] | ||
*** Hepatitis C virus (genotype 1a, isolate H) [TaxID:11108]: [https://data.glygen.org/GLYDS000344 GLYDS000344] | ***Hepatitis C virus (genotype 1b, isolate Japanese) [TaxID:11116]: [https://data.glygen.org/GLYDS000345 GLYDS000345] | ||
*** Hepatitis C virus (genotype 1b, isolate Japanese) [TaxID:11116]: [https://data.glygen.org/GLYDS000345 GLYDS000345] | ***SARS coronavirus (SARS-CoV-1) [TaxID:694009]: [https://data.glygen.org/GLYDS000467 GLYDS000467] | ||
*** SARS coronavirus (SARS-CoV-1) [TaxID:694009]: [https://data.glygen.org/GLYDS000467 GLYDS000467] | ***SARS coronavirus (SARS-CoV-2 or 2019-nCoV) [TaxID:2697049]:[https://data.glygen.org/GLYDS000434 GLYDS000434] | ||
*** SARS coronavirus (SARS-CoV-2 or 2019-nCoV) [TaxID:2697049]:[https://data.glygen.org/GLYDS000434 GLYDS000434] | *'''Glycan:''' | ||
* '''Glycan:''' | **Please see the GlyGen dataset [https://data.glygen.org/GLYDS000281 GLYDS000281] to access the current GlyGen glycan masterlist. | ||
** Please see the GlyGen dataset [https://data.glygen.org/GLYDS000281 GLYDS000281] to access the current GlyGen glycan masterlist. | |||
<div style="float: right;"> [[#top|[top]]]</div> | <div style="float: right;"> [[#top|[top]]]</div> | ||
===The reference or the source database reflects different data than what is represented in the GlyGen pages for the same annotation?=== | ===The reference or the source database reflects different data than what is represented in the GlyGen pages for the same annotation?=== | ||
* The data in GlyGen is downloaded on a specific date (“freeze date”) from the source which may also represent a specific release of that database (e.g. UniProtKB Nov. 2018 release). | *The data in GlyGen is downloaded on a specific date (“freeze date”) from the source which may also represent a specific release of that database (e.g. UniProtKB Nov. 2018 release). | ||
* If the source data changes after the GlyGen freeze date, or if there is a synchronization delay, there could be a discrepancy in both databases. | *If the source data changes after the GlyGen freeze date, or if there is a synchronization delay, there could be a discrepancy in both databases. | ||
===Does GlyGen filter out publication information on the Protein detail page?=== | ===Does GlyGen filter out publication information on the Protein detail page?=== | ||
* Yes, GlyGen filters out some publications of large scale studies that are based on genome sequencing, protein sequencing, cDNA, chromosomes that do not provide any functional annotation related to the protein. | *Yes, GlyGen filters out some publications of large scale studies that are based on genome sequencing, protein sequencing, cDNA, chromosomes that do not provide any functional annotation related to the protein. | ||
===What is ECO or eco_id (eco_identifier)?=== | ===What is ECO or eco_id (eco_identifier)?=== | ||
* The Evidence & Conclusion Ontology (ECO) is a controlled vocabulary that describes scientific evidence, which results from a variety of research methods, as well as interpretations by authors and scientific curators. ECO is used to document specific evidence to support conclusions that result from a scientific investigation. | *The Evidence & Conclusion Ontology (ECO) is a controlled vocabulary that describes scientific evidence, which results from a variety of research methods, as well as interpretations by authors and scientific curators. ECO is used to document specific evidence to support conclusions that result from a scientific investigation. | ||
* Examples: | *Examples: | ||
** Inferred from Experiment (EXP) [http://www.evidenceontology.org/term/ECO:0000269/ ECO:0000269] experimental evidence used in manual assertion. | **Inferred from Experiment (EXP) [http://www.evidenceontology.org/term/ECO:0000269/ ECO:0000269] experimental evidence used in manual assertion. | ||
** Traceable Author Statement (TAS) [http://www.evidenceontology.org/term/ECO:0000304/ ECO:0000304] | **Traceable Author Statement (TAS) [http://www.evidenceontology.org/term/ECO:0000304/ ECO:0000304] | ||
** Inferred from Sequence Ontology (ISO) [http://www.evidenceontology.org/term/ECO:0000266/ ECO:0000266] | **Inferred from Sequence Ontology (ISO) [http://www.evidenceontology.org/term/ECO:0000266/ ECO:0000266] | ||
** Inferred from Sequence or Structural Similarity (ISS) [http://www.evidenceontology.org/term/ECO:0000250/ ECO:0000250] | **Inferred from Sequence or Structural Similarity (ISS) [http://www.evidenceontology.org/term/ECO:0000250/ ECO:0000250] | ||
** Click [http://www.evidenceontology.org/browse/ here] to browse the ECO. | **Click [http://www.evidenceontology.org/browse/ here] to browse the ECO. | ||
===How to access or download all the GlyGen dataset BCOs?=== | |||
The individual BCOs can be accessed from [https://data.glygen.org/ data.glygen.org]. Use filter options to locate the required BCO and click it for detail. select the required version and the download option is on the right. | |||
The json [https://data.glygen.org/ln2releases/v-1.5.30/jsondb/bcodb.json download script] for all the GlyGen and OncoMX BCOs that are stored in MongoDB. | |||
==References== | ==References== | ||
| Line 246: | Line 277: | ||
==External links== | ==External links== | ||
* '''GlyGen.org: Informatics Resources for Glycoscience'''<br /> https://www.glygen.org/<br /> '''"Proteomics", 2012: Reference details'''<br /> http://www.unicarbkb.org/references/2844<br /> '''Protein Ontology Report - hSHBG/iso:[1/4]/SigPep-/Glyco:11''' | *'''GlyGen.org: Informatics Resources for Glycoscience'''<br /> https://www.glygen.org/<br /> '''"Proteomics", 2012: Reference details'''<br /> http://www.unicarbkb.org/references/2844<br /> '''Protein Ontology Report - hSHBG/iso:[1/4]/SigPep-/Glyco:11''' | ||
* PR:000045342 - http://purl.obolibrary.org/obo/PR_000045342 | *PR:000045342 - http://purl.obolibrary.org/obo/PR_000045342 | ||
<div style="float: right;"> [[#top|[top]]]</div> | <div style="float: right;"> [[#top|[top]]]</div> | ||
Latest revision as of 19:36, 12 February 2021
The frequently asked questions are a collection of user questions related to the GlyGen frontend, backend, and data. The answers to these questions contain definition and explanations of terms, such as Proteoform or Uniprot Accession number, and short how-to's for using the GlyGen portal and related tools, such as "How can I search all the glycosylation sites present on my protein?". The list of questions is subdivided into questions related to protein, proteoform, and general questions. You can also use the GlyGen's contact page to reach out to us with any additional questions or queries.
Protein FAQs
FAQs related to protein centric data and views.
How to Use Simple Protein Search?
- You can search for proteins by specifying their UniProtKB Accession numbers, their specific structures or the specific biochemical contexts within which they are found. For details, please refer to https://www.glygen.org/protein_search.html#tutorial
How to Use Simple Glycoprotein Search?
- You can search for glycoproteins by specifying their UniProtKB Accession numbers, their specific structures or the specific biochemical contexts within which they are found. For details, please refer to https://www.glygen.org/glycoprotein_search.html#tutorial
What does protein-centric data include?
- Protein-centric data include data types and information about a particular protein-coding gene, or that can be mapped to the canonical protein sequence representing that gene. Examples include pathways, Gene Ontology, localization, etc.
What is a UniProtKB canonical accession?
- UniProtKB canonical accession is an accession assigned to the protein isoform chosen to be the canonical sequence to which all positional annotation refers in the UniProtKB entry page.
- UniProtKB represents different isoforms of the same protein by assigning it a protein accession followed by the number of the isoform.For example: for protein accession P38398, different isoforms are represented as P38398-1, P38398-2, P38398-3, P38398-4, etc. where P38398-1 is the chosen canonical accession.
- UniProtKB uses specific criteria for choosing the canonical sequence for the entry. To know more about canonical isoforms and the canonical choosing criteria, refer to the UniProtKB help page.
How do I find a UniProtKB accession for my protein?
- UniProtKB accession is used to represent a protein by UniProtKB database. Visit UniProtKB for additional information.
- In GlyGen, you can find the UniProtKB accession for your protein either by providing a protein name, protein sequence, gene name or even by adding different cross-references (e.g. RefSeq accession, KEGG etc.):
- Go to the EXPLORE tab on GlyGen the home page and select Protein Search.
- Select Advanced Search and add the desired input by selecting the appropriate box.
How can I search all the glycosylation sites present on my protein?
- Go to the EXPLORE tab on the GlyGen home page and select Protein Search.
- Under Simple Search, select Protein from the Any category dropbox and add the UniProtKB accession.
- Click on Search.
- Click on the UniProtKB accession from the list page and navigate to the Glycosylation section.
- You can also view glycosylation sites through our highlighting feature in the Sequence section.
Proteoform FAQs
- FAQs related to proteoform and site centric data and views.
What is a proteoform?
- The term "proteoform" designates, "all of the different molecular forms in which the protein product of a single gene can be found, including changes due to genetic variations, alternatively spliced RNA transcripts and post-translational modifications," (Smith et al. Nat Methods. 2013 Mar;10(3):186-7. PMID:23443629).
- A protein can have many different glycosylated proteoforms that differ from each other in a number of ways: the site(s) that are glycosylated, the glycan structure at each modified site, and the presence of any other modifications. For example, the figure below shows several different glycosylated proteoforms of the sex hormone binding globulin (SHBG) that are represented in the Protein Ontology (PRO) .
- These forms have an OGalNAc modification at Thr36 and one of several different N4GlcNAc modifications at Asn380 or Asn396.
What does proteoform-centric data include?
- Proteoform-centric data include data types and information that is about some particular proteoform or that pertains to a specific amino acid site. A proteoform is the specific protein product of a gene; multiple proteoforms might arise due to differences in genetic variation, alternative splicing or translation start site selection, or post-translational modifications.
Why is the representation of proteoforms useful?

- Many aspects of protein function-activity, sub-cellular localization, and interaction partners, for example, are influenced by the precise combination of modifications on the protein. When proteins are represented and annotated in knowledge resources without regard to modification state, these distinctions can be lost. In contrast, proteoform-level representation makes it possible to associate annotation unambiguously with the most relevant protein form. For example, the figure below shows the relative abundances of 12 different glycosylated proteoforms of SHBG (six different glycan structures at two different sites, Asn380 and Asn396). This abundance of information is associated with the corresponding proteoform terms in Protein Ontology (PRO) .
How can we represent and proteoforms through PRO ID's when proteins have many potential glycosylation sites and glycan forms?
- The policy of the Protein Ontology (PRO) is to only represent proteoforms that have been experimentally observed. For example, if glycosylation is observed on two different sites, each of the singly modified forms can be represented in PRO; however, the proteoform with glycosylation on both sites will not be represented unless there is solid experimental evidence for its existence. In this way, the number of proteoforms per protein does not grow too large, and the forms represented are those that are of highest interest to biologists.
How can search for all proteins that bear my glycan/GlyTouCan Ac?
- Go to the Explore tab and click on Glycoprotein.
- In the Advanced Search, add your GlyTouCan accession in the Interacting Glycan field and click on Search.
- Please refer to How do I find a GlyTouCan for my glycan structure on how to find a GlyTouCan accession for your desired glycan.
Glycan FAQs
FAQs related to glycan centric data and views.
How to use simple glycan search?
- This tutorial illustrates how to search for a glycan or collection of glycans based on their general properties, structural features, attachment to a glycoprotein(s), mechanisms of biosynthesis, etc.
What does glycan-centric data include?
- Glycan-centric data include data types and information such as motifs, type/sub-type, mass, cross-references to different databases (UniCarbKB, ChEBI etc.) and sequences/string representation (e.g. IUPAC, WURCS, GlycoCT, etc.).
How do I find a GlyTouCan accession for my glycan composition or structure?
- If the glycan composition (such as type and number of monosaccharide(s)) is known:
- Visit GlyGen's composition search to search for a glycan within the GlyGen set (please note: The GlyGen glycan set captures roughly ~30% of the entire GlyTouCan collection.)
- or utilize the compo2wurcs tool developed by GlyCosmos to search against the entire GlyTouCan database.
- If the composition is not available in GlyTouCan, please refer to FAQ How can I register my glycan structure into GlyTouCan? to see how to register your glycan composition.
- If additional information (such as arrangement of monosaccharides, linkage positions, anomeric configuration, glycosidic bonds, etc) about the glycan is known:
- Visit GlyTouCan Graphic Input
- Draw the glycan structure and provide available linkage, glycosidic bonds, etc information.
- Click SEARCH
- If the drawn glycan structure is not present in GlyTouCan, please refer to the FAQ How can I register my glycan structure into GlyTouCan? on how to register your glycan.
- If a sequence/string-representation (such as GlycoCT, IUPAC extended,IUPAC condensed, KCF ,WURCS, etc) is known:
- Visit GlyCosmos Text Input
- Insert the string format to get the accession.
- To convert your sequence into a desired format, please refer to FAQ How can I convert my glycan sequence/string-format to different desired formats (e.g IUPAC, WURCS, GlycoCT, LinearCode, etc.)?
- If the GlyTouCan accession is not found, please refer to FAQ How can I register my glycan structure into GlyTouCan? on how to register your sequence.
How can I register my glycan structure into GlyTouCan?
- To register a glycan composition:
- Utilize the compo2wurcs tool developed by GlyCosmos.
- To register a glycan structure or sequence/string-representation:
- Visit the GlyTouCan website.
- Click on Registration and sign in through your desired google account.
- To draw your structure and click on Graphical Input (PubMed)
- Once the structure is complete, a sequence is generated from the input and sent to the database to check if it was previously registered. If the structure is found in the database, a link and accession number indicating the structure ID will be displayed; otherwise, a final confirmation screen will be shown. If the submit button is entered to indicate confirmation, the newly “submission ref” and graphical representation will be displayed. As all sequences will be stored in GlycoCT (condensed) format, the sequence initially input will be displayed under original structure, while the sequence converted into WURCS will be displayed under Structure.
- To register sequence/text-representation (WURCS/GlycoCT) use the Text Input.
- Please refer to FAQ 3.6 to convert your sequences into a desired format.
- To draw your structure and click on Graphical Input (PubMed)
Why do identical glycan structure images have different GlyTouCan IDs?
- Identical images (e.g. G70994MS/G29068FM or G86089ZC/G64581RP) might have different GlyTouCan accessions because the WURCS string is different. For example for
How can I convert my glycan sequence/string-format to different desired formats (e.g IUPAC, WURCS, GlycoCT, LinearCode, etc.)?
- GlycanFormatConverter (PMID:30535258) allows users to convert glycan sequences from different import formats (such as IUPAC-Condensed, IUPAC-Extended, GlycoCT, KCF, LinearCode, WURCS) to the desired export formats (such as IUPAC-Short,IUPAC-Condensed,IUPAC-Extended,GlycoCT,WURCS,GlycanWeb).
- Please click here to visit the GlycanFormatConverter GitHub repository or see the below example (IUPAC-extended to WURCS) for step-by-step instructions on using the tool:
- Go to GlycanFormatConverter Swagger and select iupacextended2wurcs
- In the import field add your IUPAC string and specify "txt"/"json" in the format field.
- Once you select Try it out, you can see the WURCS string under Response Body
How can I search all the glycans present on my protein?
- Go to the EXPLORE tab on the GlyGen home page and select Glycan Search.
- Under Simple Search, select Protein from Any category add your UniProtKB accession in the search box.
- Click on Search.
How do I input the structure of the required glycan?
You can use the GNOme (beta testing is currently underway) widget to search for your desired glycan structure or composition.
please see: https://gnome.glyomics.org/restrictions/GNOme_GlyGen.browser.html?GalNAc=1&Gal=1
By clicking on structures you can move further along glycan tree to retrieve a more specific structure (i.e with defined linkage information)
for eg: https://gnome.glyomics.org/restrictions/GNOme_GlyGen.browser.html?saccharide=G16612QG
You can right-click on your desired structure to see the glycan entry in GlyGen database, where you can retrieve additional information. (go to "Found Glycoprotein" section on the individual glycan entry page to find the associated proteins).
Alternatively, you can use the GlyGen composition search to search for your glycan with desired monosaccharides composition.
e.g.: https://www.glygen.org/glycan_list.html?id=8691cfa82cb8eea7bf6b68995ae37bc2
You can click on the desired GlyTouCan accession to retrieve more information about the glycan.
Another option is to use the GlyTouCan's graphic input tool to draw your desired glycan and retrieve the GlyTouCan accession.
Once you have the GlyTouCan accession you can use GlyGen's glycan or glycoprotein searches to find your desired output.
How is glycan data collected, integrated, and processed in GlyGen?
GlyGen processes glycoprotein-centric and glycan-centric data from different sources like GlyConnect, UniCarbKB, GlyTouCan, MatrixDB, etc in coordination between GlyGen team members at George Washington University and Georgetown University. GlyGen is also part of the GlySpace alliance and coordinates with team members biocuration activities. The overview of this process can be seen in image.
General FAQs
Does GlyGen have a tutorial on how to use the site?
- Tutorials for specific searches can be accessed through the EXPLORE tab on GlyGen home page.
What are the different resources integrated into GlyGen?
- Click here to understand which resources are integrated into GlyGen.
What licence does GlyGen use?
- GlyGen’s data is licensed under CC BY 4.0. You are free to share (copy and redistribute the material in any medium or format) and adapt (remix, transform, and build upon the material) for any purpose, even commercially as long as you give appropriate credit and attribution. To learn about CC BY 4.0, click here.
- GlyGen’s source code and data processing scripts are licenced under GNU General Public License v3.0 which gives the user freedom to use the software for any purpose, to change the software to suit the needs, to share the software with your friends and neighbors, and also the freedom to share the changes a user makes. To learn about GNU GPL V3.0, visit the GNU licenses page.
What is GlyGen's policy regarding copyright and database distribution?
- You must give appropriate credit to GlyGen (by referring to GlyGen for the whole resource or Object ID for individual datasets) and may do so in any reasonable manner, but not in any way that suggests the licencor endorses you or your use.
- Datasets made available through GlyGen include archival of versions. We recommend authors to use Creative Commons Attribution licence (CC BY) for all versions when they share their datasets with GlyGen.
- This means that users of GlyGen are entitled to use, reproduce, disseminate or display these datasets provided the original authors and GlyGen are attributed.
How can I access the GlyGen Data?
- GlyGen data can be accessed through the DATA tab on the GlyGen homepage or directly via GlyGen data page. Data can also be be accessed by querying SPARQL endpoint (coming in future) or through GlyGen Web Service API
How can I download GlyGen data?
- To download a list of results (with one or more filters) or to download data specific to an individual record:
- Go to the EXPLORE tab on the GlyGen home page and select a category for your search (e.g. protein, glycan, glycoprotein).
- Use the Simple Search option to search data based on a single filter.
- For example, select Protein from Any category to search data only by UniProtKB accession.
- Use the Advanced Search option to apply additional filters to your search.
- Click on the Search button to navigate to the list page and click the download button in the top right corner to access the results.
- To download an individual record, select a record from the list page and click the download button.
- To download the entire set of data:
- Go to the DATA tab on the GlyGen homepage.
- Select view more on the desired data object and select the download button.
- To know more about GlyGen data objects go to General FAQ What is a GlyGen data object.[top]
What is a GlyGen data object?
- GlyGen data is organized into individual data objects which are assigned a unique GlyGen ID (e.g. GLYDS000001).
- Each GlyGen data object can be accessed through the DATA tab on the GlyGen home or directly via the Glygen data page.
- Each data object has a README describing the source, contributors, integration process and quality control workflow, etc. Go to General FAQ 8 for more information.
- The data objects can be filtered based on different categories (Protein, Proteoform, Glycan), by species (Homo sapiens, Mus musculus, Rattis norvegicus, SARS-CoV-1, SARS-CoV-2 etc.), and by file type (.csv, .fasta, .png, etc.).
What is the format of the README files?
- All readme files follow the BioCompute specifications. Technical specifications can be found here .
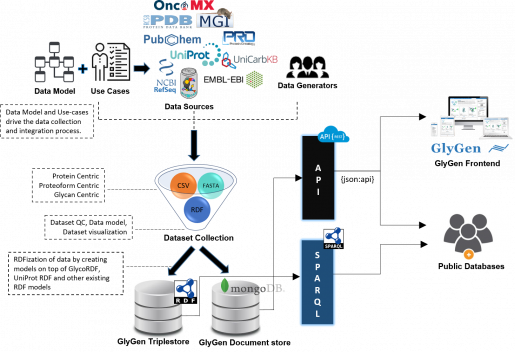
How is data integrated into GlyGen?
- Please refer to image.
How can I submit my data?
- Please use our Contact page to submit your data to GlyGen.
How often is the GlyGen data updated?
- GlyGen data is released every six months during March-April and August-September of each year.
- Minor updates to the data are made when a bug or a data issue has been found and addressed.
How can I access the previous versions of the data?
- Go to the DATA tab on the GlyGen home page.
- Click view details on the desired data object.
- Select the desired version and date from the Version dropbox on the top left corner and click on download.
Which UniProtKB and GlyTouCan accessions are inclded in GlyGen?
- GlyGen maintains a strict protein (UniProtKB) and glycan (GlyTouCan) accession list compiled based on different criteria.
- The UniProtKB accession list serves as a reference for generating protein-centric and proteoform-centric datasets, whereas the GlyTouCan accession list is used for generating glycan-centric datasets. The criteria and the statistics may be subject to change with every major and minor release.
- Protein:
- Please see the following species specific GlyGen datasets to see the current GlyGen protein masterlists:
- Homo sapiens [TaxID:9606]: GLYDS000001
- Mus musculus [TaxID:10090]: GLYDS000007
- Rattus norvegicus [TaxID:10116]:GLYDS000244
- Hepatitis C virus (genotype 1a, isolate H) [TaxID:11108]: GLYDS000344
- Hepatitis C virus (genotype 1b, isolate Japanese) [TaxID:11116]: GLYDS000345
- SARS coronavirus (SARS-CoV-1) [TaxID:694009]: GLYDS000467
- SARS coronavirus (SARS-CoV-2 or 2019-nCoV) [TaxID:2697049]:GLYDS000434
- Please see the following species specific GlyGen datasets to see the current GlyGen protein masterlists:
- Glycan:
- Please see the GlyGen dataset GLYDS000281 to access the current GlyGen glycan masterlist.
The reference or the source database reflects different data than what is represented in the GlyGen pages for the same annotation?
- The data in GlyGen is downloaded on a specific date (“freeze date”) from the source which may also represent a specific release of that database (e.g. UniProtKB Nov. 2018 release).
- If the source data changes after the GlyGen freeze date, or if there is a synchronization delay, there could be a discrepancy in both databases.
Does GlyGen filter out publication information on the Protein detail page?
- Yes, GlyGen filters out some publications of large scale studies that are based on genome sequencing, protein sequencing, cDNA, chromosomes that do not provide any functional annotation related to the protein.
What is ECO or eco_id (eco_identifier)?
- The Evidence & Conclusion Ontology (ECO) is a controlled vocabulary that describes scientific evidence, which results from a variety of research methods, as well as interpretations by authors and scientific curators. ECO is used to document specific evidence to support conclusions that result from a scientific investigation.
- Examples:
- Inferred from Experiment (EXP) ECO:0000269 experimental evidence used in manual assertion.
- Traceable Author Statement (TAS) ECO:0000304
- Inferred from Sequence Ontology (ISO) ECO:0000266
- Inferred from Sequence or Structural Similarity (ISS) ECO:0000250
- Click here to browse the ECO.
How to access or download all the GlyGen dataset BCOs?
The individual BCOs can be accessed from data.glygen.org. Use filter options to locate the required BCO and click it for detail. select the required version and the download option is on the right.
The json download script for all the GlyGen and OncoMX BCOs that are stored in MongoDB.
References
- Kahsay, R.; Vora, J.; Navelkar, R.; Mousavi, R.; Fochtman, B.; Holmes, X.; Pattabiraman, N.; Ranzinger, R.; Mahadik, R.; Williamson, T.; Kulkarni, S.; Agarwal, G.; Martin, M.; Vasudev, P.; García Castro, L.; Edwards, N.; Zhang, W.; Natale, D.; Ross, K.; Mazumder, R. (2020)."GlyGen data model and processing workflow". Bioinformatics. btaa238. https://doi.org/10.1093/bioinformatics/btaa238
- York, W. S., Mazumder, R., Ranzinger, R., Edwards, N., Kahsay, R., Aoki-Kinoshita, K. F., Campbell, M. P., Cummings, R. D., Feizi, T., Martin, M., Natale, D. A., Packer, N. H., Woods, R. J., Agarwal, G., Arpinar, S., Bhat, S., Blake, J., Castro, L., Fochtman, B., Gildersleeve, J., … Zhang, W. (2020). GlyGen: Computational and Informatics Resources for Glycoscience. Glycobiology, 30(2), 72–73. PMID: 31616925 https://doi.org/10.1093/glycob/cwz080
- Sumer-Bayraktar, Z., Nguyen-Khuong, T., Jayo, R., Chen, D. D., Ali, S., Packer, N. H., & Thaysen-Andersen, M. (2012). Micro- and macroheterogeneity of N-glycosylation yields size and charge isoforms of human sex hormone binding globulin circulating in serum. Proteomics, 12(22), 3315–3327. https://doi.org/10.1002/pmic.201200354 PMID: 23001782 https://pubmed.ncbi.nlm.nih.gov/23001782/
External links
- GlyGen.org: Informatics Resources for Glycoscience
https://www.glygen.org/
"Proteomics", 2012: Reference details
http://www.unicarbkb.org/references/2844
Protein Ontology Report - hSHBG/iso:[1/4]/SigPep-/Glyco:11 - PR:000045342 - http://purl.obolibrary.org/obo/PR_000045342