Protein details/Glycosylation: Difference between revisions
| (7 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
The | The Glycosylation section of the [[Protein details]] page in GlyGen provides a detailed list of glycosylation sites, reported glycans attached to the protein, and sites text mined from publications. | ||
==Glycosylation== | ==Glycosylation== | ||
[[File:Protein_Glycosylation_Screenshot.jpg|thumb|upright|Screenshot of the Glycosylation section on the [[Protein | [[File:Protein_Glycosylation_Screenshot.jpg|thumb|upright|Screenshot of the Glycosylation section on the [[Protein details]] page in GlyGen.]] | ||
Glycosylation is presented in 4 tabs on the [[Protein | Glycosylation is presented in 4 tabs on the [[Protein details]] page in GlyGen. The Glycosylation summary provides the overall information about the section like the total number of sites (O,N,S,C linked sites), total number of N linked and O-linked sites with the total number of glycan annotations (structures) observed on the protein. Eg. 18 site(s) total, 169 N-linked annotation(s) at 17 site(s), 1 O-linked annotation(s) at 1 site(s) | ||
===Reported Sites with Glycan=== | ===Reported Sites with Glycan=== | ||
| Line 18: | Line 18: | ||
This tab shows reported glycosylation sites without glycan information. The following columns are presented in the table: | This tab shows reported glycosylation sites without glycan information. The following columns are presented in the table: | ||
* '''Source''' - GlyGen evidence linking to the databases and papers that provided the glycosylation information | *'''Source''' - GlyGen evidence linking to the databases and papers that provided the glycosylation information | ||
* '''Type''' - Type of glycosylation. Eg. N-linked, O-linked | *'''Type''' - Type of glycosylation. Eg. N-linked, O-linked | ||
* '''Residue''' - Amino acid residue of the given protein along with its position. Eg. Asn294 | *'''Residue''' - Amino acid residue of the given protein along with its position. Eg. Asn294 | ||
* '''Note''' - Additional information about the entry such as curation notes, O-glycosylation subtype, remarks, etc. | *'''Note''' - Additional information about the entry such as curation notes, O-glycosylation subtype, remarks, etc. | ||
=== Predicted Only === | ===Predicted Only=== | ||
This tab shows predicted glycosylation sites. The following columns are presented in the table: | This tab shows predicted glycosylation sites. The following columns are presented in the table: | ||
=== Text Mining === | {{Expand section|small=no}} | ||
===Text Mining=== | |||
{{Expand section|small=no}} | |||
<br /> | <br /> | ||
==Source of information== | ==Source of information== | ||
The Glycosylation data is collected and integrated from the resources such as '''[https://www.uniprot.org/ UniProtKB]''', [https://glyconnect.expasy.org/ '''Glyconnect'''], '''[http://www.unicarbkb.org/ UniCarbKB], [https://www.rcsb.org/ RCSB PDB], [https://www.oglcnac.mcw.edu/ The O-GlcNAc Database]'''. | The Glycosylation data is collected and integrated from the resources such as '''[https://www.uniprot.org/ UniProtKB]''', [https://glyconnect.expasy.org/ '''Glyconnect'''], '''[http://www.unicarbkb.org/ UniCarbKB], [https://www.rcsb.org/ RCSB PDB], [https://www.oglcnac.mcw.edu/ The O-GlcNAc Database]'''. | ||
*[https://www.uniprot.org/ UniProtKB] - only reported sites information and predicted information is downloaded from UniProtKB | *[https://www.uniprot.org/ '''UniProtKB'''] - only reported sites information and predicted information is downloaded from UniProtKB | ||
*[https://glyconnect.expasy.org/ Glyconnect] - reported sites with glycan information on known and unknown residues is downloaded from Glyconnect | *[https://glyconnect.expasy.org/ '''Glyconnect'''] - reported sites with glycan information on known and unknown residues is downloaded from Glyconnect | ||
*UniCarbKB - reported sites with glycan information on known and unknown residues is downloaded from UniCarbKB | *'''[http://unicarbkb.org/ UniCarbKB]''' - reported sites with glycan information on known and unknown residues is downloaded from UniCarbKB | ||
*[https://www.rcsb.org/ RCSB PDB] - only reported sites information is downloaded from RCSB PDB | *[https://www.rcsb.org/ '''RCSB PDB'''] - only reported sites information is downloaded from RCSB PDB | ||
*[https://www.oglcnac.mcw.edu/ The O-GlcNAc Database] - Only O-GlcNAcylation reported sites with glycan information on known and unknown residues is downloaded from The O-GlcNAc database | *[https://www.oglcnac.mcw.edu/ '''The O-GlcNAc Database'''] - Only O-GlcNAcylation reported sites with glycan information on known and unknown residues is downloaded from The O-GlcNAc database | ||
==Data access== | ==Data access== | ||
The collected data is processed and stored in '''[https://data.glygen.org data.glygen.org]''' in the following datasets. | |||
The collected data is processed and stored in '''[https://data.glygen.org data.glygen.org]''' in following datasets. | |||
Homo Sapiens (Human) Datasets | Homo Sapiens (Human) Datasets | ||
Latest revision as of 20:16, 19 May 2022
The Glycosylation section of the Protein details page in GlyGen provides a detailed list of glycosylation sites, reported glycans attached to the protein, and sites text mined from publications.
Glycosylation
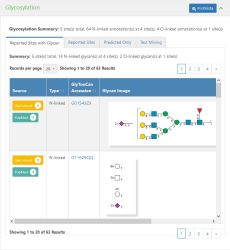
Glycosylation is presented in 4 tabs on the Protein details page in GlyGen. The Glycosylation summary provides the overall information about the section like the total number of sites (O,N,S,C linked sites), total number of N linked and O-linked sites with the total number of glycan annotations (structures) observed on the protein. Eg. 18 site(s) total, 169 N-linked annotation(s) at 17 site(s), 1 O-linked annotation(s) at 1 site(s)
Reported Sites with Glycan
This tab shows reported glycosylation sites with glycan information. The following columns are presented in the table:
- Source - GlyGen evidence linking to the databases and papers that provided the glycosylation information
- Type - Type of glycosylation. Eg. N-linked, O-linked
- GlyTouCan ID - Unique accession assigned to the registered glycan structure in GlyTouCan database. Eg. G01543ZX
- Glycan Image - Image of the glycan in SNFG format
- Residue - Amino acid residue of the given protein along with its position. Eg. Asn294
- Note - Additional information about the entry such as curation notes, O-glycosylation subtype, remarks, etc.
Reported Sites
This tab shows reported glycosylation sites without glycan information. The following columns are presented in the table:
- Source - GlyGen evidence linking to the databases and papers that provided the glycosylation information
- Type - Type of glycosylation. Eg. N-linked, O-linked
- Residue - Amino acid residue of the given protein along with its position. Eg. Asn294
- Note - Additional information about the entry such as curation notes, O-glycosylation subtype, remarks, etc.
Predicted Only
This tab shows predicted glycosylation sites. The following columns are presented in the table:
This section needs expansion. You can help by adding to it. |
Text Mining
This section needs expansion. You can help by adding to it. |
Source of information
The Glycosylation data is collected and integrated from the resources such as UniProtKB, Glyconnect, UniCarbKB, RCSB PDB, The O-GlcNAc Database.
- UniProtKB - only reported sites information and predicted information is downloaded from UniProtKB
- Glyconnect - reported sites with glycan information on known and unknown residues is downloaded from Glyconnect
- UniCarbKB - reported sites with glycan information on known and unknown residues is downloaded from UniCarbKB
- RCSB PDB - only reported sites information is downloaded from RCSB PDB
- The O-GlcNAc Database - Only O-GlcNAcylation reported sites with glycan information on known and unknown residues is downloaded from The O-GlcNAc database
Data access
The collected data is processed and stored in data.glygen.org in the following datasets.
Homo Sapiens (Human) Datasets
- Human Glycosylation Sites (UniProtKB; GLY_000038)
- Glycosylation Sites (UniCarbKB [Human proteins]; GLY_000040)
- Human Glycosylation Sites (RCSB PDB; GLY_000042)
- Human Glycosylation Sites (GlyConnect; GLY_000329)
- Human Glycosylation Sites ([GPTwiki]; GLY_000480)
- Human Glycosylation Sites ([Automatic Literature Mining] [Automatically verified]; GLY_000481)
- Human O-GlcNAc Glycosylation Sites (MCW; GLY_000517)
- Human Glycosylation Sites UniCarbKB Glycomics Study (GLY_000611)
Hepatitis C Virus Datasets
- HCV1a Glycosylation Sites (Literature + UniCarbKB; GLY_000335)
- HCV1a Glycosylation Sites (UniProtKB; GLY_000382)
- HCV1b Glycosylation Sites (UniProtKB; GLY_000383)
SARS Coronavirus Datasets
- SARS-CoV2 Glycosylation Sites (UniProtKB; GLY_000473)
- Glycosylation Sites (UniCarbKB [SARS CoV 2 proteins]; GLY_000479)
- SARS-CoV1 Glycosylation Sites (UniProtKB; GLY_000495)
- SARS-CoV2 Glycosylation sites (UniprotKB; GLY_000473)
- SARS-CoV1 Glycosylation Sites (Literature; GLY_000612)
Mus musculus (Mouse) Datasets
- Mouse Glycosylation Sites (UniProtKB; GLY_000039)
- Glycosylation sites (UniCarbKB [Mouse proteins]; GLY_000041)
- Mouse Glycosylation Sites (RCSB PDB; GLY_000043)
- Mouse Glycosylation Sites (GlyConnect; GLY_000330)
Rattus norvegicus (Rat) Datasets
- Glycosylation sites (UniCarbKB [Rat proteins]; GLY_000221)
- Rat Glycosylation Sites (UniProtKB; GLY_000224)
- Rat Glycosylation Sites (RCSB PDB; GLY_000226)
- Rat Glycosylation Sites (GlyConnect; GLY_000331)
Data harmonization
This section needs expansion. You can help by adding to it. |
Data filtering
This section needs expansion. You can help by adding to it. |